How to use Digital Walnut
1. About Digital Walnut
2. The data of DW database
3. How to browse the DW data?
4. Genome browser
5. How to search gene?
6. How to download?
1. About Digital Walnut
Digital Walnut (DW), a comprehensive, fast-growing database, provides a one-stop resource for users to browse, search and download data for researches of Juglandaceae plants transparently. It will play important role in studies of origin and evolution, formation of traits, selection of parents in crossbreeding and oriented genetic improvement of cultivars on walnut and other woody plants.
2. The data of DW database
Digital Walnut housed the most comprehensive high-throughput sequencing data from 13 species and 3 interspecific hybrids.
We collected, screened, and integrated 16 sets of whole genome assembly data and their corresponding annotation information, 322 sets of annotated and searchable reference transcriptomes, combining peer-reviewed published RNA-seq as well as EST datasets, protein sequences, and miRNAs, and one set each of epigenomics and rhizosphere microbial genome data.
3. How to browse the DW data?
Users can browse all data in Digital Walnut database via three tabs.
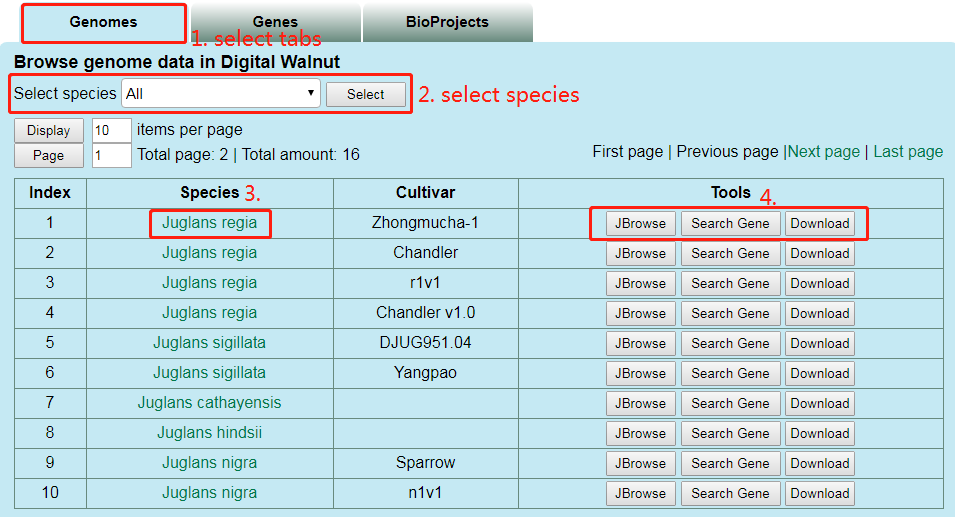
(1) browse data via genomes
Users can select species at part 2 and click the link at part 3 to jump to the introduction page of species. At part 4, users can click the corresponding button to go to the Genome Browse page, search page and download page.
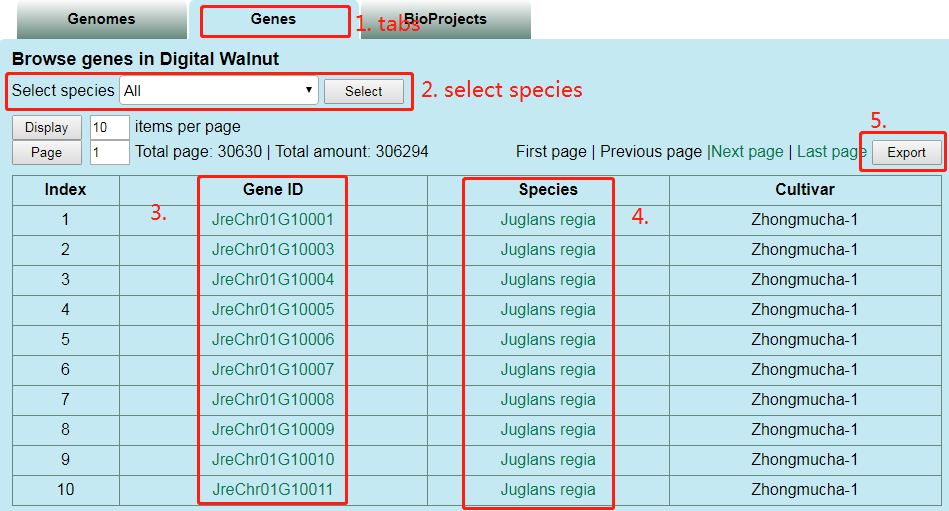
(2) browse data via genes
Users can select species at part 2, click the link at part 3 to jump to the detail information of gene, click the link at part 4 to jump to the introduction page of species and click ‘Export’ button at part 5 to get the result list.
(3) browse data via bioprojects
Users can select bioproject at part 2, click the link at part 3 to jump to the introduction page of species, click the link at part 4 to jump to the corresponding project page in NCBI, click the link at part 5 to jump to the article and click ‘Export’ button at part 6 to get the result list.

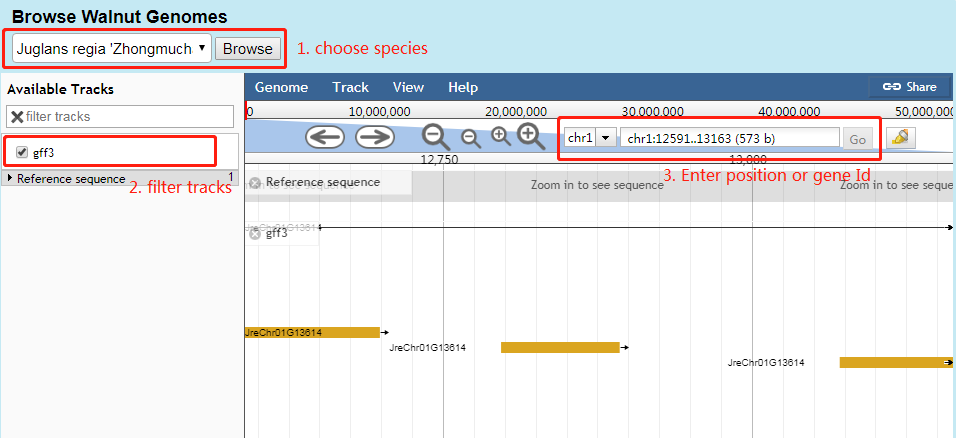
4. Genome browser
Users can click Genome Browser on Navigation Bar to jump to genome browser webpage to see genome and annotation information of species.
Users can select species at part 1, tracks at part 2, postion or gene Id at part 3. The genome annotation are shown.
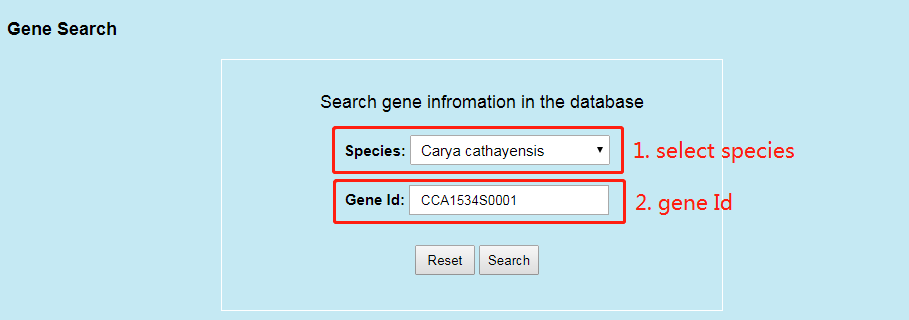
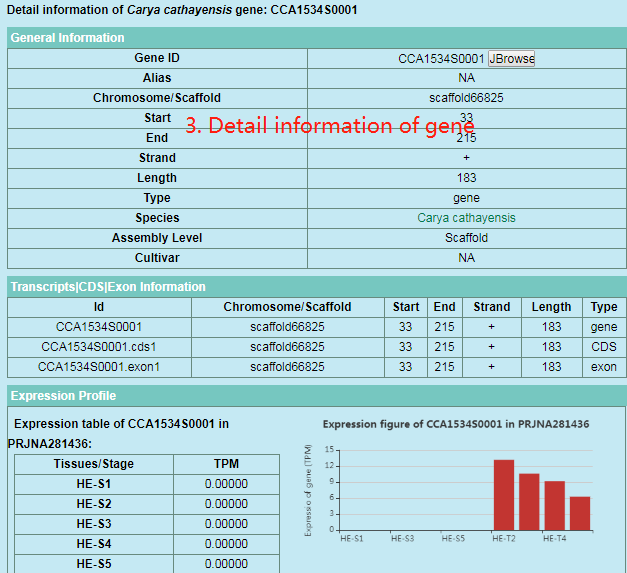
5. How to search gene?
Users can search the gene information in Search webpage via gene ID.
Users can select species at part 1 and enter gene Id at part 2 and then click ‘Search’ button to get the detail information of gene at part 3.
6. How to download?
Users can get all data in DW database on Downloads webpage. Users can click their interested datasets and begin to download. Also, there are download/Export buttons in browse.