Help
Search in NPInter database
In most of the NPInter pages, there is a search bar on the top.

The search bar can be filled in with molecular name or molecular IDs (NONCODE, miRBase, UniProt, circBase).
Browse in NPInter database
All interactions can be browsed and searched in the browse module.
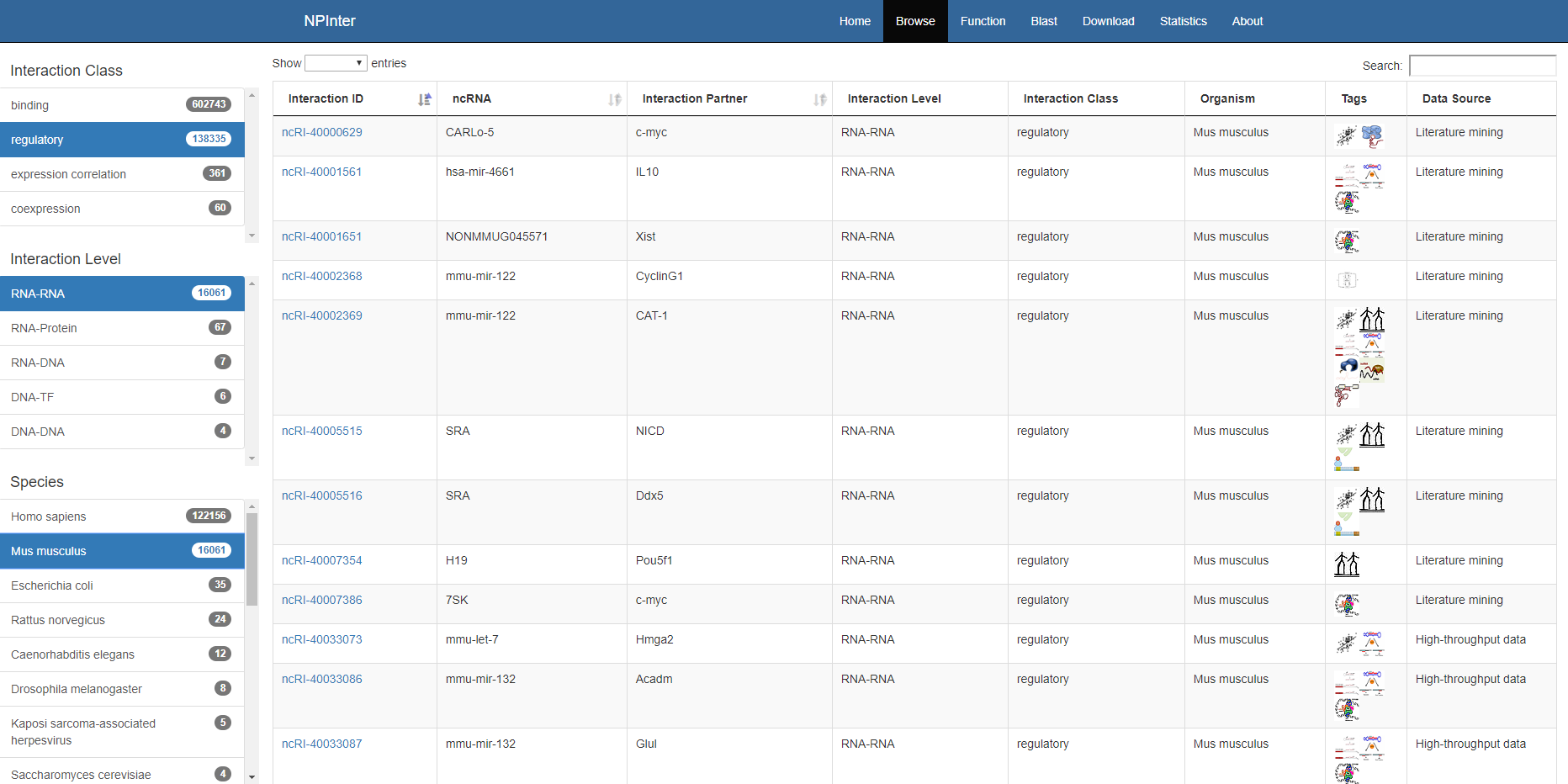
User can filter the interactions with interaction class, interaction level, species, data source and tissue through the filter panel on the left.

After filtering by these attributes, users can also search molecule name in the search bar on the top of the data table.

Enjoy browsing!
Browse and search RBPs
In NPInter V5.0, we built the RBP module to provide the interactions of RNA-binding proteins (RBPs) with annotations of localization, binding domains and functions.

User can filter the RBPs with different properties through the filter panel on the left or just click on the buttons in the table. The following filtering conditions means filter RBPs with Essential Genes or Splicing regulation function and Nuclei localization and Helicase domain.
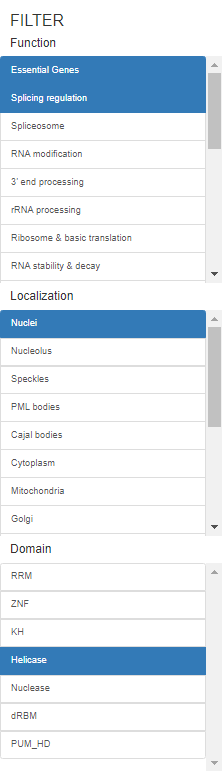
Users can also search the name or Ensembl ID of a certain RBP in the search bar on the top of the data table.

Click on the RBP name in the data table, user can jump to the molecular page of the RBP with detailed interaction information (eg: ADAR ).
Hope this module will provide a useful and convenient resource portal for the study of the RNA-RBP network.
Download data
All interaction data can be freely downloaded from the Download page.
We provide both the whole dataset of NPInter v4.0 and v3.0.

Disease search
User can search interactions and molecules related to certain disease.
The disease search is on the Function page.
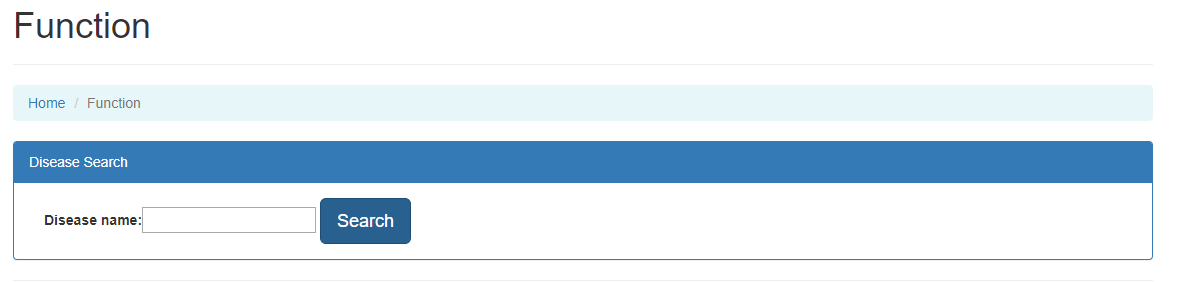
In this page, users can enter the disease name and search for molecules related to this disease.

Blast sequence
In NPInter blast module, we support searching by nucleotide sequence. Users can either update a fasta file or fill in the form with the whole sequence in FASTA format.
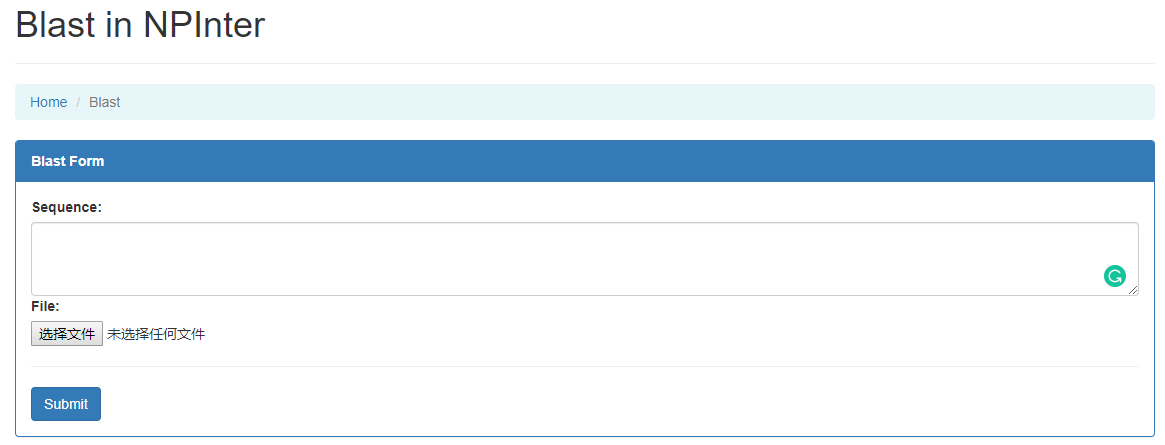
There is a result sample

Users can click the Details button to enter the molecule profile page for corresponding molecule.