Help
Search MEIs
How to search MEIs
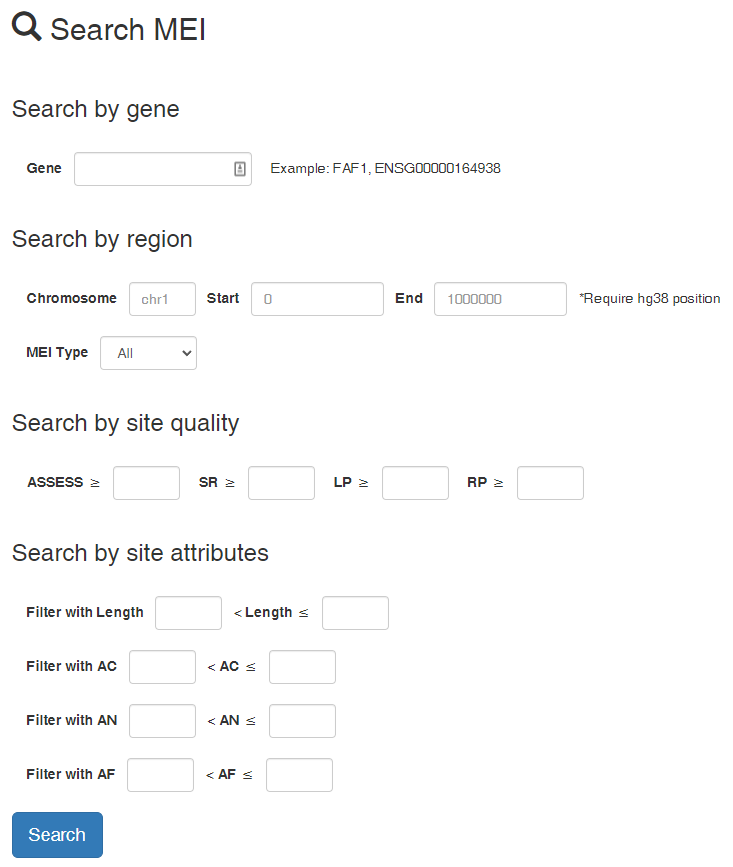
MEIs can be searched in the Home page. We provide three types of search module. The filters in the three modules can be used simultaneously.
- Search by gene: HGNC gene symbol and Ensembl Gene Stable ID can be used
- Search by region: Region can be specified (If gene name has been entered, the gene coordinates will be used) and MEI Type can be chosen
- Search by site quality: MEI sets can be filtered by ASSESS, SR, LP, and RP.
- Search by site attributes: MEI sets can be filtered by site length, AC, AN and AF.
About site quality
| Attribute | Description |
|---|---|
| ASSESS | Provides information on evidence availible to decide insertion site. 0 = No overlapping reads at site; 1 = Imprecise breakpoint due to greater than expected distance between evidence; 2 = discordant pair evidence only -- No split read information; 3 = left side TSD evidence only; 4 = right side TSD evidence only; 5 = TSD decided with split reads, highest possible quality |
| SR | Number of split reads |
| LP | Total number of discordant pairs supporting the left side of the breakpoint |
| RP | Total number of discordant pairs supporting the right side of the breakpoint |
About the search result
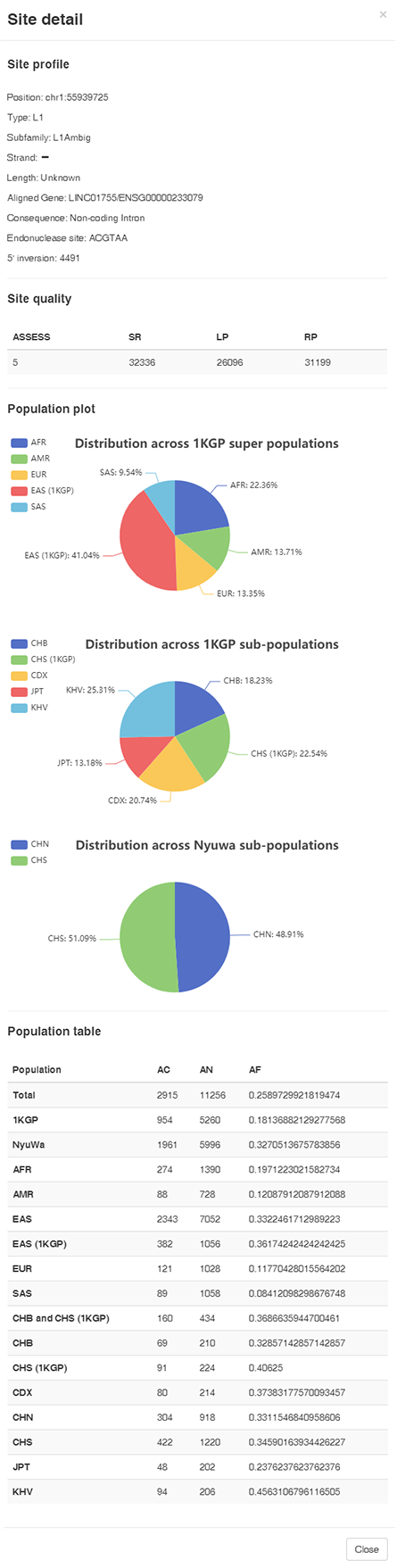
Each entry in the result table links to a Site Detail page. The page shows the profile of the MEI including the position, MEI type, subfamily, aligned gene, consequence, GWAS SNP LD region overlapped, quality attributes, and frequency in different populations.
Site profile
- Position: The mobile element insert site position
- Type: The MEI Type (Alu, L1, SVA, HERVK)
- Subfamily: Subfamily predicted by MELT (only for Alu and L1)
- Strand: The strand of the MEI
- Length: The length of the MEI. When the length can't be detected, the field is "unknown"
- Aligned gene: If the MEI is located in gene region, this field provide the gene symbol and Ensembl stable ID
- Consequence: The functional annotation of the MEI aligned region (Intergenic, Enhancer, CDS, Coding intron, Noncoding Intron, Noncoding Exon, UTR, flanking sequence)
- Endonuclease site: The endonuclease site beside the MEI
- 5' inversion: The 5' inversion position of L1 insertions. The position is from the 3' end (The same as the MELT ISTP field).
- GWAS SNP LD: We extracted GWAS SNPs (p-value < 1e-8) from the GWAS catelog database. The LD region was predicted by LD SNPs in 1KGP samples. This field provides the associated SNPs which have LD with MEI site.
Site quality
| Attribute | Description |
|---|---|
| ASSESS | Provides information on evidence availible to decide insertion site. 0 = No overlapping reads at site; 1 = Imprecise breakpoint due to greater than expected distance between evidence; 2 = discordant pair evidence only -- No split read information; 3 = left side TSD evidence only; 4 = right side TSD evidence only; 5 = TSD decided with split reads, highest possible quality |
| SR | Number of split reads |
| LP | Total number of discordant pairs supporting the left side of the breakpoint |
| RP | Total number of discordant pairs supporting the right side of the breakpoint |
Population table
- Projects: Samples in our study are from 1KGP and Nyuwa projects
- Subpopulation:
- The population names of 1KGP projects are the same as the original classification.
- The population in Nyuwa were devided into two subpopulations: CHN (North Han Chinese), and CHS (South Han Chinese)